Note
Click here to download the full example code
E step, Kalman filter¶
Inner working of the E step GLE_analysisEM.GLE_Estimator
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/examples/plot_e_step.py:17: FutureWarning: Passing a negative integer is deprecated in version 1.0 and will not be supported in future version. Instead, use None to not limit the column width.
pd.set_option("display.max_colwidth", -1)
{'A': array([[ 0.02645208, 1. ],
[-1. , 1.6849652 ]]), 'C': array([[ 1.00000000e+00, -1.67919914e-17],
[-3.58404070e-17, 1.00000000e+00]]), 'force': array([[-1.]]), 'µ_0': array([0.]), 'Σ_0': array([[1.]]), 'SST': array([[0.00026452, 0. ],
[0. , 0.01684965]]), 'dt': 0.005, 'basis': {'basis_type': 'linear', 'transformer': None, 'dim_x': 1, 'nb_basis_elt': 1, 'mean': array([0.]), 'n_output_features': 1}}
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:103: RuntimeWarning: Mean of empty slice.
hSdouble = 0.5 * np.log(detd[detd > 0.0]).mean()
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/conda/latest/lib/python3.9/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
/home/docs/checkouts/readthedocs.org/user_builds/gle-analysisem/checkouts/latest/GLE_analysisEM/_gle_estimator.py:104: RuntimeWarning: Mean of empty slice.
hSsimple = 0.5 * np.log(dets[dets > 0.0]).mean()
{'A': array([[ 0.02645208, 1. ],
[-1. , 1.6849652 ]]), 'C': array([[ 1.00000000e+00, -2.65775124e-16],
[-4.93096709e-17, 1.00000000e+00]]), 'force': array([[-1.]]), 'µ_0': array([0.]), 'Σ_0': array([[1.]]), 'SST': array([[ 2.64520814e-04, -1.73472348e-18],
[-1.73472348e-18, 1.68496520e-02]]), 'dt': 0.005, 'basis': {'basis_type': 'linear', 'transformer': None, 'dim_x': 1, 'nb_basis_elt': 1, 'mean': array([0.]), 'n_output_features': 1}}
Real datas
dxdx [[0.0003105327689501738, 3.421232830534903e-05], [3.421232830534903e-05, 0.016813548574053382]]
xdx [[-0.00011558038291017877, 0.004690654455607655], [-0.0047642304638938135, -0.008419940568312167]]
xx [[0.9011063718703859, -0.025579194692135276], [-0.025579194692135276, 0.9535644807589472]]
bkx [[0.013926371314893363, 0.004575700354467006]]
bkdx [[-0.00453191153991419, 0.00011368340416723901]]
bkbk [[0.8948908969005819]]
µ_0 [-0.20328789892473598]
Σ_0 [[0.0]]
hS NaN
dtype: object
Estimated datas
dxdx [[0.0003105327689501738, 4.139405031848609e-05], [4.139405031848609e-05, 0.016939558734364833]]
xdx [[-0.00011558038291017877, 0.0047719838764412525], [-0.004819320300032244, -0.00847955320308491]]
xx [[0.9011063718703859, -0.016194579896374562], [-0.016194579896374562, 0.9589237375785946]]
bkx [[0.013926371314893363, 0.010360234927923786]]
bkdx [[-0.00453191153991419, 4.9370791776712645e-05]]
bkbk [[0.8948908969005819]]
µ_0 [0.2904700508604896]
Σ_0 [[0.3378154504392095]]
hS -0.638995
dtype: object
Diff
dxdx [[0.0, 0.20991620181588788], [0.20991620181588788, 0.00749456069647959]]
xdx [[0.0, 0.01733860841878234], [-0.01156321814318897, -0.007079935337915734]]
xx [[0.0, 0.3668846853355458], [0.3668846853355458, 0.005620235367179342]]
bkx [[0.0, 1.2641856164837486]]
bkdx [[0.0, -0.5657168067901666]]
bkbk [[0.0]]
µ_0 [2.428860509636294]
Σ_0 [[inf]]
hS NaN
dtype: object
[[ 0.00014039 0.00497411]
[-0.00513833 0.008756 ]]
[[2.63865309e-04 1.32586614e-07]
[1.32586614e-07 1.68407917e-02]] [[0.00026452 0. ]
[0. 0.01684965]]
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from GLE_analysisEM import GLE_Estimator, GLE_BasisTransform, sufficient_stats, sufficient_stats_hidden
# Printing options
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
pd.set_option("display.width", None)
pd.set_option("display.max_colwidth", -1)
dim_x = 1
dim_h = 1
random_state = None
force = -np.identity(dim_x)
A = [[5, 1.0], [-1.0, 2.07]]
C = np.identity(dim_x + dim_h) #
basis = GLE_BasisTransform(basis_type="linear")
generator = GLE_Estimator(verbose=1, dim_x=dim_x, dim_h=dim_h, basis=basis, C_init=C, force_init=force, init_params="random", random_state=random_state)
X, idx, Xh = generator.sample(n_samples=10000, n_trajs=10, x0=0.0, v0=0.0)
traj_list_h = np.split(Xh, idx)
time = np.split(X, idx)[0][:, 0]
for n, traj in enumerate(traj_list_h):
traj_list_h[n] = traj_list_h[n][:-1, :]
print(generator.get_coefficients())
est = GLE_Estimator(init_params="user", dim_x=dim_x, dim_h=dim_h, basis=basis)
est.set_init_coeffs(generator.get_coefficients())
est.dt = time[1] - time[0]
est._check_initial_parameters()
Xproc, idx = est.model_class.preprocessingTraj(est.basis, X, idx_trajs=idx)
traj_list = np.split(Xproc, idx)
est.dim_coeffs_force = est.basis.nb_basis_elt_
datas = 0.0
for n, traj in enumerate(traj_list):
datas_visible = sufficient_stats(traj, est.dim_x)
zero_sig = np.zeros((len(traj), 2 * est.dim_h, 2 * est.dim_h))
muh = np.hstack((np.roll(traj_list_h[n], -1, axis=0), traj_list_h[n]))
datas += sufficient_stats_hidden(muh, zero_sig, traj, datas_visible, est.dim_x, est.dim_h, est.dim_coeffs_force) / len(traj_list)
est._initialize_parameters(None)
print(est.get_coefficients())
print("Real datas")
print(datas)
new_stat = 0.0
noise_corr = 0.0
for n, traj in enumerate(traj_list):
datas_visible = sufficient_stats(traj, est.dim_x)
muh, Sigh = est._e_step(traj) # Compute hidden variable distribution
new_stat += sufficient_stats_hidden(muh, Sigh, traj, datas_visible, est.dim_x, est.dim_h, est.dim_coeffs_force) / len(traj_list)
print("Estimated datas")
print(new_stat)
print("Diff")
print((new_stat - datas) / np.abs(datas))
Pf = np.zeros((dim_x + dim_h, dim_x))
Pf[:dim_x, :dim_x] = 5e-3 * np.identity(dim_x)
YX = new_stat["xdx"].T - np.matmul(Pf, np.matmul(force, new_stat["bkx"]))
XX = new_stat["xx"]
A = -np.matmul(YX, np.linalg.inv(XX))
Pf = np.zeros((dim_x + dim_h, dim_x))
Pf[:dim_x, :dim_x] = 5e-3 * np.identity(dim_x)
# A = generator.friction_coeffs
print(A)
bkbk = np.matmul(Pf, np.matmul(np.matmul(force, np.matmul(new_stat["bkbk"], force.T)), Pf.T))
bkdx = np.matmul(Pf, np.matmul(force, new_stat["bkdx"]))
bkx = np.matmul(Pf, np.matmul(force, new_stat["bkx"]))
residuals = new_stat["dxdx"] + np.matmul(A, new_stat["xdx"]) + np.matmul(A, new_stat["xdx"]).T - bkdx.T - bkdx
residuals += np.matmul(A, np.matmul(new_stat["xx"], A.T)) - np.matmul(A, bkx.T) - np.matmul(A, bkx.T).T + bkbk
print(residuals, generator.diffusion_coeffs)
# SST = 0.5 * (residuals + residuals.T)
fig, axs = plt.subplots(1, dim_h)
# plt.show()
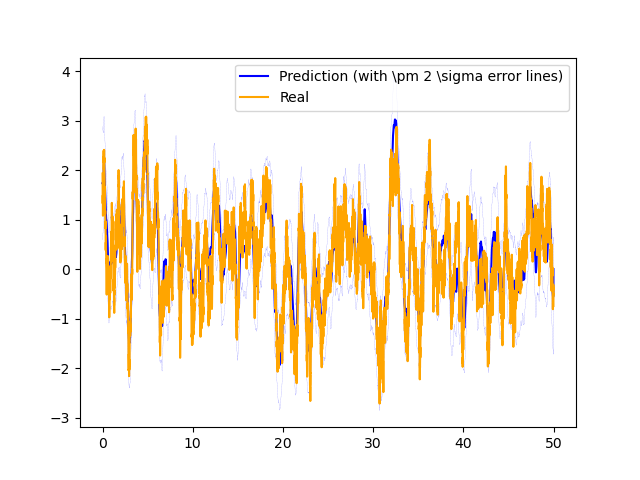
for k in range(dim_h):
axs.plot(time[:-1], muh[:, k], label="Prediction (with \\pm 2 \\sigma error lines)", color="blue")
axs.plot(time[:-1], muh[:, k] + 2 * np.sqrt(Sigh[:, k, k]), "--", color="blue", linewidth=0.1)
axs.plot(time[:-1], muh[:, k] - 2 * np.sqrt(Sigh[:, k, k]), "--", color="blue", linewidth=0.1)
axs.plot(time[:-1], traj_list_h[n][:, k], label="Real", color="orange")
axs.legend(loc="upper right")
plt.show()
Total running time of the script: ( 0 minutes 18.367 seconds)